A simple strain typing assay for Trypanosoma cruzi: discrimination of major evolutionary lineages from a single amplification product
Abstract
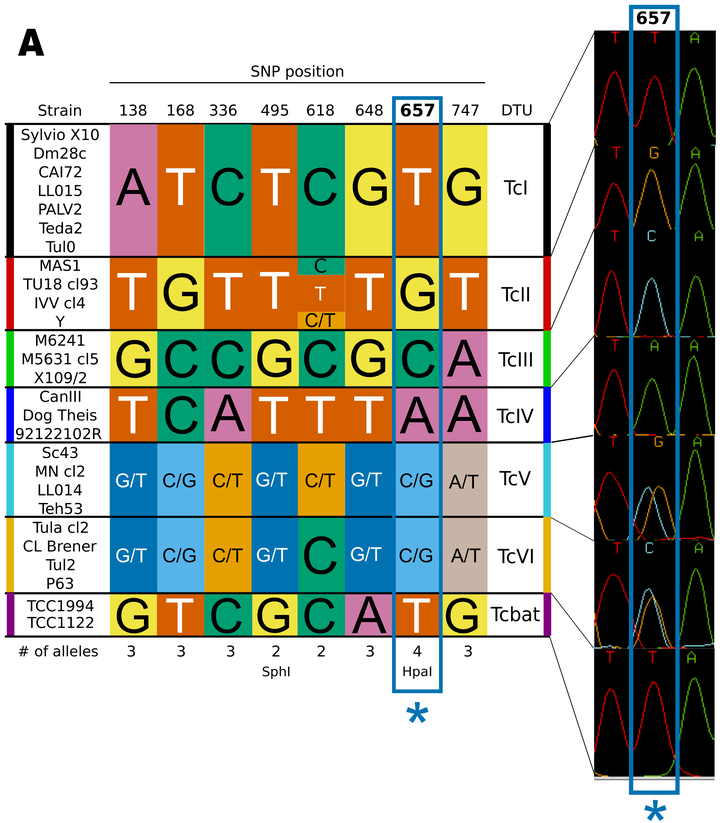
Trypanosoma cruzi, the causative agent of Chagas Disease, infects approximately 8 million people in the Americas, with 200,000 new cases reported anually. The disease, in its chronic stage, has different manifestations: mega-colon, mega-esophagus, and cardiomyopathy, among others. The fact that infections by the same species cause these different clinical outcomes is believed to be determined, at least in part, by the genetic background of the parasite (infection by different strains). By analyzing a number of molecular markers, the population of the parasite has been divided into seven major evolutionary lineages, which evolve mostly independently, by clonal expansion with infrequent exchange of genetic material. Accurate classification of different strains and isolates into their corresponding evolutionary lineages is therefore essential to obtain a good map of biological, biochemical and ecoepidemiological features for the whole species. The current methods available to type T. cruzi stocks are either laborious and costly (requiring the amplification and sequencing of a variable number of genes or gene fragments), or limited in resolution. In this work we describe a number of key discriminant sites in a gene encoding a putative enzyme from the sterol pathway of the parasite, which were exploited to design a couple of alternative typing assays. Using these key discriminant sites, we can classify any T. cruzi stock into either six or seven evolutionary lineages using only one gene fragment, and in a matter of hours (depending on the assay used). To our knowledge, the proposed assays are the first typing assays that can discriminate T. cruzi stocks with such speed and low cost.