Alanine Scan for known Chagas antigens, November 2019 update
High-throughput mutational analysis of Trypanosoma cruzi antigenic epitopes reveal consistent conservation of key residues across human Chagas Disease populations.
 Logo ANPCyT
Logo ANPCyT
High-throughput mutational analysis of Trypanosoma cruzi antigenic epitopes reveal consistent conservation of key residues across human Chagas Disease populations.
Abstract
Chagas Disease is a major health problem for which no vaccine for public health interventions are yet available. Diagnosis is essential for the identification of infection and ecoepidemiological surveillance. Diagnosis is routinely performed using serological methods, which require well characterized antigens. Although available tests give satisfactory results in most cases, discordant results remain as a possible cause of undetected infected patients. We have previously conducted a large-scale screening of T. cruzi linear B-cell epitopes using high-density peptide arrays, leading to the development of a new proof-of-principle multiepitope diagnostic test that has excellent diagnostic performance1. However, understanding which residues in an epitope are important for antibody binding can lead to improved reagents. To further characterize known antigens, we performed Alanine scans of 649 different proteins (881 antibody-binding peaks/epitopes, represented by 2,913 peptides). This experiment was based on replacing each amino acid residue in each peptide for an Alanine (or a Glycine if the original residue was itself an Alanine), and assessing the impact on reactivity of each modified epitope. Using this peptide array design we have assayed 45,492 peptide variants against 108 Chagas Disease serum samples (from 6 different countries). We developed an algorithm to integrate and analyze the effect of these epitope mutations, and to visualize key residues for each antigen and sample. We identified precise residue positions in epitopes that play a fundamental role in the seroreactivity. In summary, we observe an average of ~6 key residues per epitope. As an example Ag2/CA-2, a known antigen2, displays the same 7 key residues in all reactive sera; In contrast, SAPA, Ag36, and B12 display different degrees of reactivity conservation of their key residues. This variable responses for different antigens, can be used in the design of improved antigens for diagnostic applications.
Introduction
When the first antigens for Chagas disease were discovered in the 80s’, there was only scarce information on the sequences recognized by antibodies1. Further epitope characterizations, employing genetic and immunochemical tools, allowed the identification and mapping of some reactive regions2. This led to improved diagnosis methods based on either recombinant proteins or fragments (e.g. SAPA) and more recently to the development of chimeric antigens composed of small antigenic determinants from disparate parasite proteins. Recently it has been possible to achieve very high throughput and performance in immune assays using high-density peptide arrays. These arrays allow massive characterizations of antigens, identifying concrete linear sequences recognized by chronic, acute and congenital Chagas patients mapped to specific regions of T. cruzi proteome3. In our laboratory, the complete T. cruzi proteome was analyzed, leading to the identification of new/novel antigens, and the simultaneous mapping of their epitopes. However, besides the delineation of the antigenic region in a protein of interest (the sequence recognized by the antibodies), little is known about the key particular residues for each epitope.
Serum selection and sectorized arrays design strategy
108 serum samples from Chagas Positive subjects were selected from the following 6 regions/countries (Argentina, Bolivia, Brazil, Colombia, Mexico and the southwestern USA as can be seen in the next summary). Sectorized microarrays were used to assay 12 individual samples in parallel per slide. In this setup, the same set of 400,000 peptides is used in each of the 12 sectors. In this way, we have been able to assay every serum sample against the entire design using 9 identical microarrays.
**Summary** for sera selection.
Serums | 108 Regions | 6 Serums per microarray | 12 Microarrays | 9
Alanine Scan
This experiment is based on permuting an alanine (or a Glycine if the original residue was itself an Alanine) in each position of the sequence, one residue at a time, and then to assess the antibody-binding to the mutated peptide. In turn, this is made for every possible peptide from the original protein sequence. Alanine is one of the smallest amino acids and, therefore, it is expected to introduce the smallest impact due to its own presence. Hence, observed changes in signal are assumed to be due to the perturbation of the functional epitope site. Hence, this scanning strategy allow us to estimate the importance of each residue in the studied sequences. In this work we analyzed 881 antibody-binding regions in selected protein sequences (“signal peaks” derived from microarray experiments). Each of these regions had a length of 20 residues with known reactivity in previous experiments with Chronic Chagas patients (mostly positive controls). For each of these 881 peaks, we performed sequential and complete mutations (Ala-scans) of 5 peptides of 15 residues length, overlapped over 14 residues (1 residue offset). In total we analyzed the antibody binding of chronic chagas disease sera to ~ 3000 original peptides.and we generated the single-Ala-mutated sequence for each residue, which resulted in ~45,000 new (mutated) peptides to assay.

Residue analysis
In order to identify the relevance of each amino acid from the studied protein sequences, we calculated the mean signal of every non-mutated peptide containing a particular amino acid (this is the original signal) and next we calculated the corresponding mutated signal as the average of every peptide with this particular residue mutated to an alanine. The resulting difference between these two signals is the Change produced by mutating this residue. This procedure allowed to quantify the importance of each residue in the antibody/epitope binding. This quantification will be essential, for example, to study if different sera recognize the same epitope in the same way (similar or identical key residues) or if there are different patterns of human antibody binding to the same epitope in different individuals.
Microarray design
In a previous work we have evaluated the entire Trypanosoma cruzi proteome in the form of short peptides against 14 different pooled samples from Chagas Disease patients from geographically diverse locations. 6 of these serum pools were from chronic chagas disease patients, and therefore we could characterize which peptides/antigenic regions/antigens were specifically recognized by chronic chagas patients sera. We have selected reactive peptides from this massive screening and have produced a second array design in which we can assay 12 individual serum samples per slide. This new design uses sectorized arrays that display 400,000 immobilized peptides per sector. In each sector, the entire Alanine Scan design is contained in ~45,000 peptides, about 10% of the entire design.
Some results as example
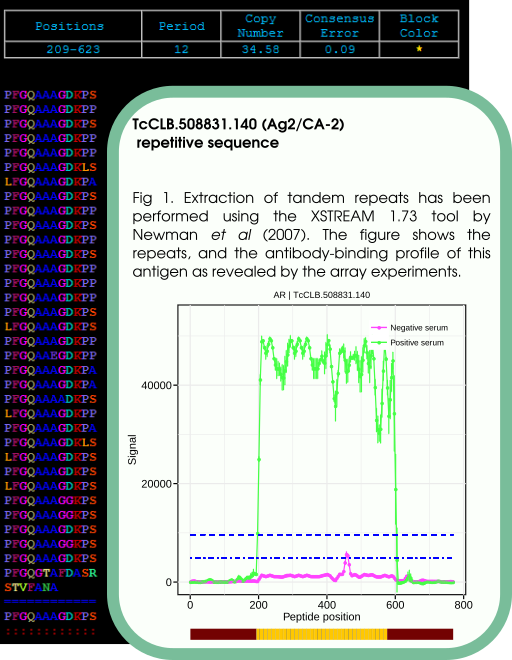
TcCLB.508831.140 (Ag2/CA-2)
This known antigen 1 displayed sero-reactivity with most of the sera (100/108). When a detailed residue analysis was performed, the same key residues were found in all serums with only small variations, as can be seen in fig. 2. However, if we focus on those serum samples with low or null reactivity, we found that some of them display the same pattern of key residues observed for highly reactive samples, suggesting that these low reactivity sera were actually recognizing this antigen but with low intensity.
TcCLB.508831.140 (Ag2/CA-2) Ala-Scan key residues (heatmap)
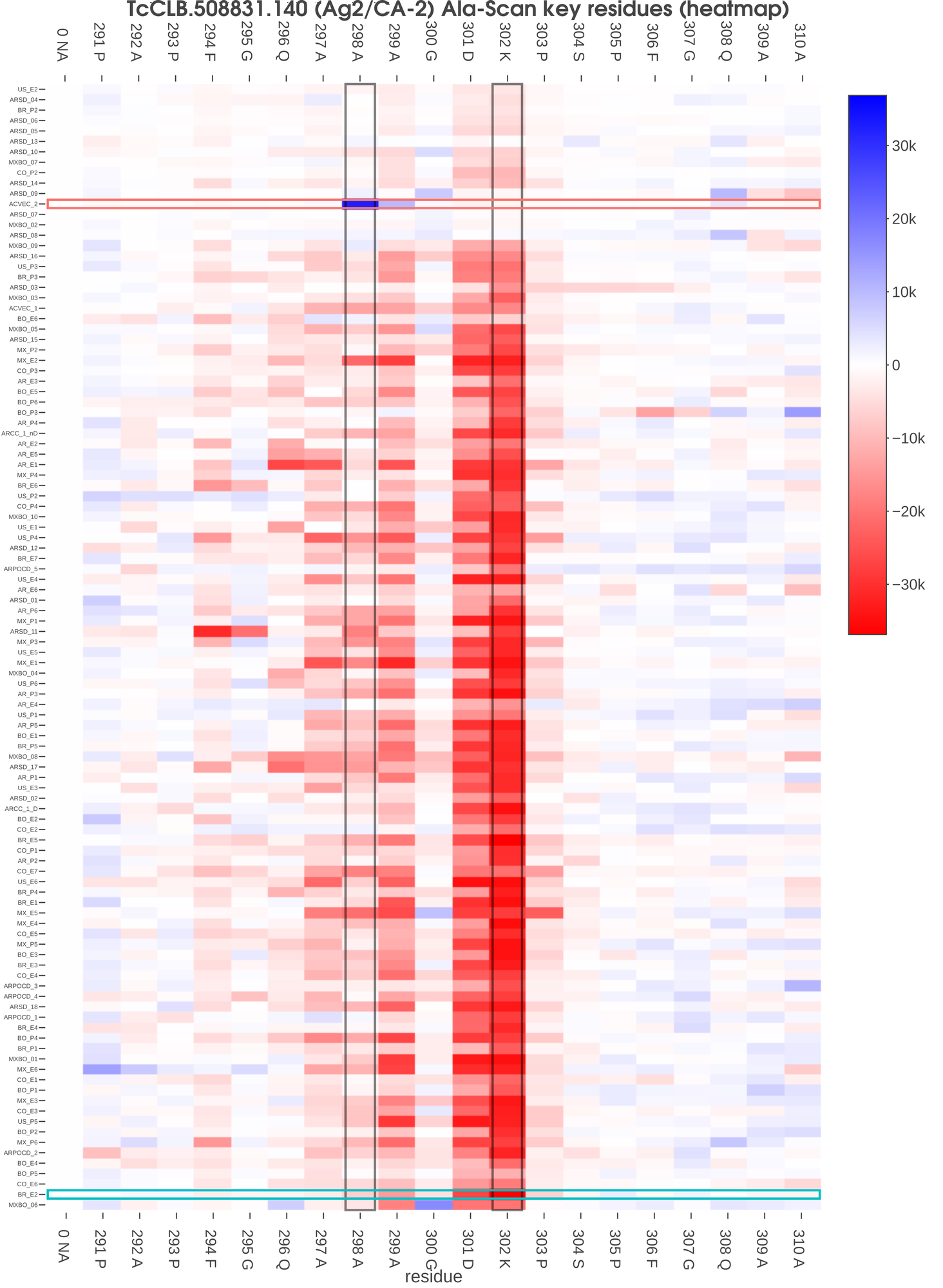
Fig 2. This heatmap visualization shows the specific signal change in each analyzed residue in all serum samples assayed. Each row represents an individual serum and each column represents a specific epitope residue. Serum samples were ordered by signal and residues by the position in the original protein. The color of each cell corresponds to the difference in the mean signal of the original peptides versus novel peptides with that particular residue mutated for an alanine (or glycine). In this case, we have found that for Ag2, most sera showed the same key residues (294 F, 297 A, 298A, 299 A, 301 D, 302 K and 303 P). In the figure we highlight two sera (ACVEC_2 and BR_E2) for which we present the corresponding protein profiles in Fig. 3. Also, we highlight two residues for which we present a scatterplot with all mutated/original peptides in Fig. 4.
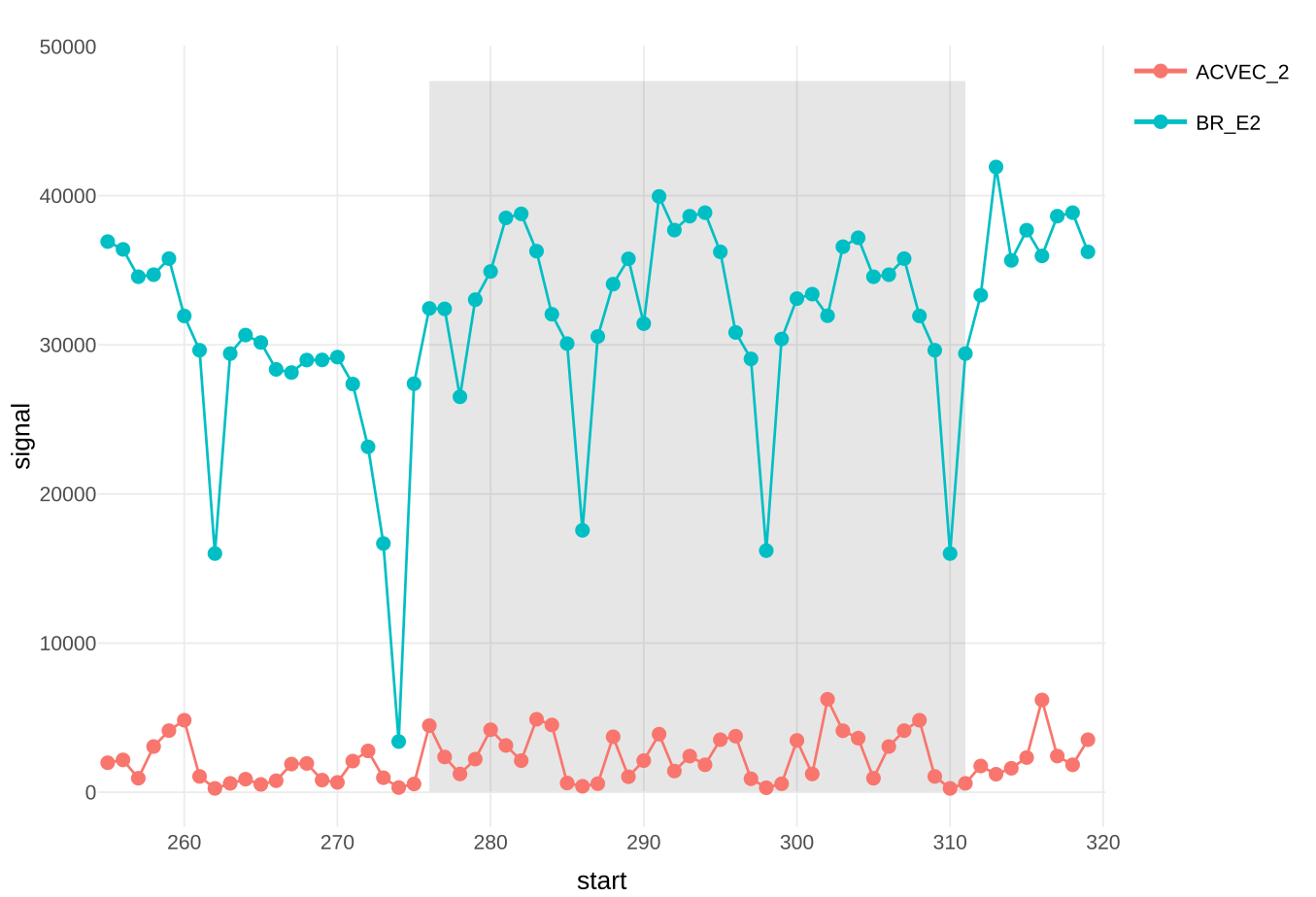
Fig 3. Protein profiles for selected sera (see Fig. 2), the X axis represent the peptide start position in the protein sequence and the Y axis presents the mean signal of both technical replicates. The area of the profile that contains the studied residues is shaded in grey.
Fig 4. All peptides needed for the estimation of the cells selected in Fig. 2 are represented in this figure (also those from technical replicates). We have separated in the plot those peptides with the original sequence from those that contain a mutated alanine or glycine.
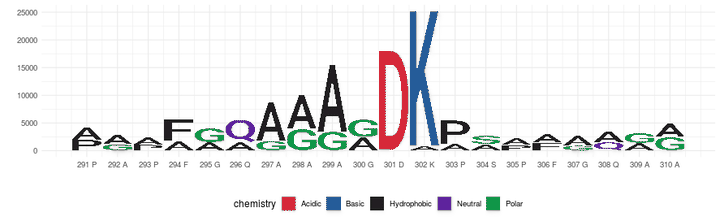
Fig 5. A sequence logo has been produced using the same information shown in Fig. 2. The height of the original amino acid is represented by the mean change for those sera with negative change, and the height of the mutated amino acid in the same position is represented by the mean change for those sera with positive change. This way, if a residue change has a negative impact on the reactivity of many sera, it is shown as relevant in the sequence logo, and at the same time, if replacing the same residue has a positive impact in the reactivity of sera, the replaced amino acid is also shown as relevant.
Bibliographie
- Ibañez, C. F., Affranchino, J. L. & Frasch, A. C. C. Antigenic determinants of Trypanosoma cruzi defined by cloning of parasite DNA. Mol. Biochem. Parasitol. 25, 175–184 (1987).
- da Silveira, J. F., Umezawa, E. S. & Luquetti, A. O. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 17, 286–291 (2001).
- Carmona, S. J. et al. Towards High-throughput Immunomics for Infectious Diseases: Use of Next-generation Peptide Microarrays for Rapid Discovery and Mapping of Antigenic Determinants. Mol. Cell. Proteomics 14, 1871–1884 (2015).