Serodiscordance and borderline serology, November 2019 update
Investigating serodiscordance and borderline serology in Chagas Disease using high-density peptide arrays.
 T. cruzi and other pathogens immune response
T. cruzi and other pathogens immune response
Investigating serodiscordance and borderline serology in Chagas Disease using high-density peptide arrays.
Abstract
Diagnosis of chronic Chagas Disease is currently based on serological techniques. Although available diagnostic tests give satisfactory results in most cases, there is currently no gold standard and discordant results remain a possible cause of undetected cases. Here, using state of the art high-density peptide arrays we examined the global human antibody repertoire of chronic Chagas Disease patients as well as serodiscordant and borderline serology cases. First, peptide arrays displaying 2.8 million unique peptides from the complete proteomes of T. cruzi strains CL-Brener (DTU TcVI, 19,668 proteins) and Sylvio X10 (DTU TcI, 10,832 proteins) were assayed with serum samples from infected subjects from Argentina, Bolivia, Brazil, Colombia, Mexico and the US, as well as negative samples from the same regions. This allowed us to identify the antigenic subset of T. cruzi peptides recognized by a diverse collection of sera. Next, in a second screening focusing on 400,000 peptides selected from this subset we assayed individual serum to study 28 serodiscordant cases from Argentina and Mexico with borderline serology. These displayed a wide reactivity based on the number of positive peptides and quantification of signal. We observed almost complete lack of correlation between the quantitative values obtained in commercial ELISA (Wiener v4.0) and those obtained by the array. Hence, there is much room for improvement of current serological diagnosis. Based on the reactivity against 6 known antigens, we have separated these sera in three groups: one group of 17 sera reactive against several antigens; a second group of 8 sera reactive with fewer antigens and a group of 3 sera that were negative against most known antigens assayed. Using this information we will shortlist novel antigens from the high-content screening to improve existing diagnostic kits. In this presentation we will revisit the concept of serodiscordance in the light of all new data arising from this screen.
Study conception
There is no gold standard for Chagas disease diagnosis and current clinical diagnosis relies on two or more serological techniques. Discordant and/or borderline results can be consequence of several factors, one of them could be the incorrect or insufficient eleccion of T. cruzi antigens. Taking advantage of the already set up and optimization -in our lab- of the technique of high density peptide microarrays for T cruzi entire proteome, we have consider analyse serum samples from serodiscordant and borderline individuals.
High density peptide microarrays
See more
Serological techniques such as ELISA has the potential of high sensibility and reproducibility. However, the process to characterization and purification for each antigen is long, expensive and tedious. With the irruption and optimization of high throughput next generation sequence it has been possible to sequenciate and publicate the entire genome for T. cruzi. Using this information, it is possible to predict every possible protein or peptide within the limits of the genome annotation technique errors. High Density peptide microarrays is a technique that allows the simultaneous assay of about 3-4 million unique short peptides. In a previous work, we have assayed every possible short peptide that can be obtain from T. cruzi predicted proteome against 72 CD patients pooled sera and non-CD pooled sera, from 6 different regions, which allowed us to determinate which peptides where reactive only against CD patients. However, this has been performed with pooled sera, because the technique is very costous for each assay. After a prioritization analysis we have elaborate a microarray design with only antigenic peptides, with only about 300.000 peptides that should take every serological marker of CD and we have produce a final microarray design with this peptides duplicated in each of the 12 sectors. As the technique has the possibility of assay in parallel 12 sera samples in each of this sectors, we have been able to assay individually 12 sera at the same time per microarray and evaluate every possible antigenic peptide, as those peptides that were not reactive against pooled sera, probably are not now against individual sera. A summary for this microarray peptide design is shown in Table 1
Antigenic sequences selected for the study
This study exploit the results from ~400.000 peptides from T. cruzi with known antigenic properties, novel ones and other from distinct pathogens that could infect humans.
Known antigens from Trypanosoma cruzi.
See more
As mentioned before, this experience arrose information from about 10000 proteins in almost 400000 peptides. Analyze and interpret data of such size is complicated, and can consume a time and computational resources even at imposible levels. In extencion, most of this antigens are novel ones, and part of this work consist of characterized them and how they react in Chronic, serodiscordant and borderline groups, with almost no previous information for this two last groups. Therefore we have start the analysis using only a fraction of antigens with reported antigenic properties as shown in Table 1 and Table X.Other pathogens peptides
See more
As this group of serodiscordant patients could have co-infections with other organism than T.cruzi that could explain -at some point- the complications in diagnosis we have added to the design a set of peptides from: Trypanosoma rangeli, Leishmania sp, Toxoplasma gondii, Treponema pallidum, Human alphaherpesvirus, Human Cytomegalovirus (HCMV), Human immunodeficiency viruses (HIV), Dengue, Chikungunya and Zika as summarized in Table 1. As the viruses proteomes are relative small, they have been entirely included. For other pathogens like T. gondii, T. pallidum, T rangeli only a subset of proteins have been included.Study design and samples
For this retrospective study we have assayed sera from 18 patients with previous serodiscordant diagnosis for CD from Argentina and 10 with borderline diagnosis from Mexico. As well, 72 chronic Chagas disease patients have been assayed against the same microarray design. Table 2 presents a summary for all sera included, and more details for serodiscordant and borderline are summarized in Table 3.
Each of these sera has been analysed, with a technical replicate, against the entire microarray design as detailed before, and following the protocol of [CITAR]. The final signal for each peptide-serum reactivity is the mean raw signal of the technical replicates. No further normalization or signal processing has been considered in this study.
**Table 1:** Microarray peptides composition by organism source.
| Total unique peptides | 392791 | ||
|---|---|---|---|
| proteins | peptides | ||
| T. cruzi Sylvio X10 and CL Brener | 9862 | 320168 | |
| T. cruzi other strains* | 2544 | 81213 | |
| Other pathogens: | 206 | 12437 | |
| Human alphaherpesvirus | 78 | 4698 | |
| HCMVM | 62 | 4080 | |
| Trypanosoma rangeli | 24 | 1224 | |
| Toxoplasma gondii | 22 | 1110 | |
| Dengue | 1* | 472 | |
| Leishmania | 7 | 419 | |
| HIV | 2 | 124 | |
| Chikungunya | 1* | 120 | |
| Zika | 1* | 103 | |
| Treponema pallidum | 3 | 87 |
*For this viruses the entire predicted proteome has been included as one single protein.
**Table 2:** Patients summary for source and previous diagnosis
| Serodiscordant | Borderline | Chronic | ||
|---|---|---|---|---|
| Argentina | 18 | 0 | 20 | |
| Bolivia | 0 | 0 | 12 | |
| Brazil | 0 | 0 | 12 | |
| Colombia | 0 | 0 | 11 | |
| México | 0 | 10 | 12 | |
| United States | 0 | 0 | 12 | |
| Total patients | 117 | |||
| Total Chronic Chagas disease | 79 | |||
| Total serodiscordant | 18 | |||
| **Total borderline | 10 |
**Table 3:** Serodiscordants and borderline summary for Chagas disease previous diagnosis.
| Serodiscordant | |||||||
|---|---|---|---|---|---|---|---|
| Sample | KIT ELISA | DO | ELISA | HAI | IFI | ||
| ARSD_01 | WIENER | NA | P | N | P | ||
| ARSD_02 | WIENER | NA | N | P | P | ||
| ARSD_03 | WIENER | NA | P | N | N | ||
| ARSD_04 | WIENER | NA | P | N | N | ||
| ARSD_05 | WIENER | NA | P | N | N | ||
| ARSD_06 | WIENER | NA | P | N | N | ||
| ARSD_07 | WIENER | NA | N | P | N | ||
| ARSD_08 | LEMOS | 0,091 | N | P | P | ||
| ARSD_09 | LEMOS | 0,129 | N | P | P | ||
| ARSD_10 | LEMOS | 0,108 | N | P | N | ||
| ARSD_11 | LEMOS | 0,174 | N | P | P | ||
| ARSD_12 | LEMOS | 0,115 | N | P | P | ||
| ARSD_13 | LEMOS | 0,066 | N | P | P | ||
| ARSD_14 | LEMOS | 0,309 | P | N | P | ||
| ARSD_15 | LEMOS | 0,428 | P | N | P | ||
| ARSD_16 | LEMOS | 0,18 | N | P | P | ||
| ARSD_17 | LEMOS | 0,163 | N | P | P | ||
| ARSD_18 | WIENER | 2,185 | P | N | P | ||
| Borderline | |||||||
| Sample | KIT ELISA | DO | ELISA | HAI | IFI | ||
| MXBO_01 | WIENER | 0,639 | P | NA | NA | ||
| MXBO_02 | WIENER | 0,261 | P | NA | NA | ||
| MXBO_03 | WIENER | 0,339 | P | NA | NA | ||
| MXBO_04 | WIENER | 0,672 | P | NA | NA | ||
| MXBO_05 | WIENER | 0,406 | P | NA | NA | ||
| MXBO_06 | WIENER | 0,536 | P | NA | NA | ||
| MXBO_07 | WIENER | 0,141 | N | NA | NA | ||
| MXBO_08 | WIENER | 0,397 | P | NA | NA | ||
| MXBO_09 | WIENER | 0,387 | P | NA | NA | ||
| MXBO_10 | WIENER | 0,267 | P | NA | NA |
WIENER refers to Chagatest ELISA recombinante 4.0, Wiener Lab, Argentina and LEMOS to BIOZIMA Chagas recombinante, laboratorio LEMOS S.R.L., Argentina.
Results
Figure 1: Known Chagas antigens and other pathogens reactivity in chronic Chagas and healthy pooled samples
Fig 1. Y axis represent the 99 percentile of the signal from peptides of known Chagas antigens (Trypanosoma cruzi) and other pathogens (Trypanosoma rangeli, Leishmania sp, Toxoplasma gondii, Treponema pallidum, Human alphaherpesvirus, Human Cytomegalovirus (HCMV), Human immunodeficiency viruses (HIV), Dengue, Chikungunya and Zika) each one in a different color. The X axis represent each pool of samples (Argentina = AR, Bolivia = BO, Brazil = BR, Colombia = CO, México = MX, United States = US) from chronic Chagas patients (PO, represent by +) or healthy individuals (NE, represent by -).
Figure 2: Known Chagas antigens and other pathogens reactivity in every individual sample (Chronic, serodiscordant, borderline) and pooled samples
Fig2. Y axis represent the 99 percentile of the signal from peptides of known Chagas antigens (Trypanosoma cruzi) and other pathogens (Trypanosoma rangeli, Leishmania sp, Toxoplasma gondii, Treponema pallidum, Human alphaherpesvirus, Human Cytomegalovirus (HCMV), Human immunodeficiency viruses (HIV), Dengue, Chikungunya and Zika) each one in a different color. The X axis represent pooled and individual samples. Chronic Chagas disease pooled samples are represented by (+) symbol, healthy pooled individuals by (-), individual samples from Chronic Chagas patients are simple circles, and serodiscordant or borderline individuals have double circles.
Figure 3: Known Chagas and other novel antigens reactivity in every individual sample (Chronic, serodiscordant, borderline) and pooled samples
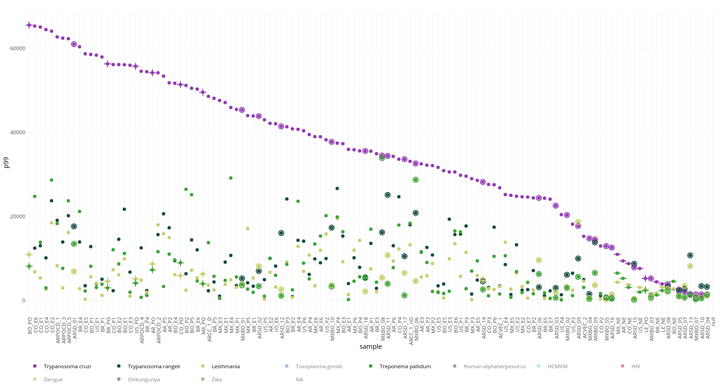
Figure 4: Known Chagas and other novel antigens reactivity in every individual sample (Chronic, serodiscordant, borderline) and pooled samples ordered by signal
Fig 4 Y axis represent the 99 percentile of the signal from peptides of known Chagas antigens (All known) and other novel antigens each one in different color. The X axis represent pooled and individual samples ordered by reactivity in each antigen. Chronic Chagas disease pooled samples are represented by (+) symbol, healthy pooled individuals by (-), individual samples from Chronic Chagas patients are simple circles, and serodiscordant or borderline individuals have double circles.
Figure 5: Known Chagas and other novel antigens best peptide reactivity in every individual sample (Chronic, serodiscordant, borderline) and pooled samples ordered by signal
Fig 5. Y axis represent the 99 percentile of the signal from only the best peptide of known Chagas antigens (All known) and other known and novel antigens, each one in different color. The X axis represent pooled and individual samples. Chronic Chagas disease pooled samples are represented by (+) symbol, healthy pooled individuals by (-), individual samples from Chronic Chagas patients are simple circles, and serodiscordant or borderline individuals have double circles.